Problems in causal modeling of transcriptional regulation
[grn Earlier, I discussed opportunities and ambitions in modeling the human transcriptome and its protein and DNA counterparts. I also promised a discussion of the many statistical issues that arise. To begin that discussion, I am trying to identify statistical issues and position them into some type of structure.
- Technological limitations
- Major aspects of cell state are unobserved
- We can’t yet scale both perturbations and readouts
- Genomics data are notoriously noisy
- Perfect measurements aren’t enough
Technological limitations
Major aspects of cell state are unobserved
In a typical single-cell RNA-sequencing experiment, we measure gene activity by sampling RNA transcripts, reading them out, and counting them. This glosses over many aspects of cell state.
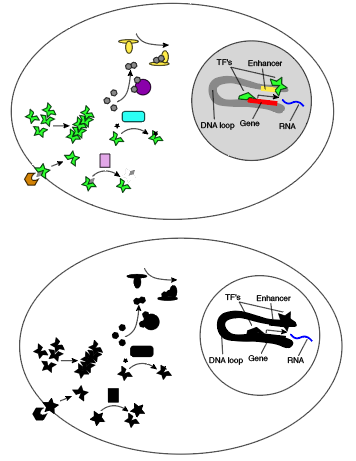
Modern technology does let us observe protein quantities and modifications, chromatin state and looping, small RNA’s, and images or phenotypes, but these are rarely coupled. You get RNA in one sample, chromatin state in another sample, proteomics in another, and spatial or phenotypic information in another. Algorithmic advances claim to allow coupling of independent single cell RNA-seq and ATAC-seq measurements on similar samples (ref 8), and modern genomics pioneer Aviv Regev envisions scRNA data as a universal plug between different types of measurements in this review, but I can’t vouch for the performance of these schemes. Even if we will eventually measure everything, we are not there yet, and so I discuss GRN modeling with missing “layers” here.
We can’t yet scale both perturbations and readouts
Large-scale perturbations cannot be coupled with genome-wide readouts. Traditional CRISPR screens are limited to single-channel readouts, and technologies such as CROP-seq are limited to dozens of perturbations. Even expensive, highly multiplexed versions of such screens are limited to ~6,000 low-impact perturbations (Gasperini et al 2019), with no information on chromatin, cell morphology, etc.
This is a technological limitation circa 2019, not a fundamental limit of any sort. There are even a couple of exciting exceptions. One of them, done with a cheap, custom measurement technology, is CMAP; it covers thousands of perturbations with a kinda-sorta-transcriptome-wide readout. Other technologies are beginning to enable genome-wide screens with single-cell imaging (Feldman et al 2019) or cheap targeted sequencing, which is an exciting direction.
Genomics data are notoriously noisy
Most technologies suffer from measurement errors to varying degrees, but I want to take this opportunity to complain to the internet about two technologies in particular that are especially messy.
- Droplet-based single-cell RNA-seq. Mammalian cells contain roughly one million transcripts per cell. They are phyically downsampled to thousands or tens of thousands during the capture process. Cell-to-cell variability is dominated by this measurement error, so single-cell RNA-seq is ironically not well suited to study subtle biological differences from cell to cell. You can average cells together in order to learn about differences in gene activity, or you can pool information across genes in order to classify cells. But you usually can’t take the data literally and say “gene X is higher in cell C1 than it is in cell C2” because the sampling error is too harsh.
- Another extreme example is the assay for transposase-accessible chromatin. It is popular due to the ease of use, and it has been adapted for single-cell assays too. But its competitor DNase-seq produces clear, distinctive signals surrounding binding sites of transcription factors: there will be a spike in DNase sensitivity on either side of the binding site, with a trough in the site itself. ATAC-seq only provides a messier version of this pattern, and consequently, it is not as useful to detect where transcription factors are binding on the genome. There is a direct comparison between the two technologies here.
Calviello, A. K., Hirsekorn, A., Wurmus, R., Yusuf, D., & Ohler, U. (2019). Reproducible inference of transcription factor footprints in ATAC-seq and DNase-seq datasets using protocol-specific bias modeling. Genome biology, 20(1), 42.
Measurement noise is too often ignored in GRN modeling. For instance, the entire exposition of the well-known package ARACNE seems to assume biological variability with informative correlations among genes. The underlying mathematics, such as the data processing equality, do not apply when measurement error is added, especially when the noise variance differs from gene to gene. Kasper Hansen, Stephanie Hicks, & Yi Wang notice a similar issue here.
Fundamental limitations
Perfect measurements aren’t enough
Even in an ideal scenario with perfect measurement, causal inference is not always possible. One of my favorite papers completely removes measurement error and missing data from consideration. It uses mathematics and simulations to consider a scenario with perfect measurements of all relevant chemicals. This is one example of the fascinating issue of non-identifiability: multiple models can produce the same exact predictions. This problem can appear in different ways:
- It’s hard to tell whether A affects B, B affects A, or C affects A and B. Time-series measurements and assays related to direct binding can help infer the direction of a connection, but a lot of people I’ve spoken with believe the field is not ready to tackle this directionality problem.
- If A affects B affects C, it’s hard to tell that A doesn’t affect C directly. This interfaces confusingly with the “missing layers” problem: if B is not measured, maybe you actually do want your model to output
A->C. - If effects are complicated and synergistic, it’s hard to tell whether A, B, C, D, and E together physically determine the regulation of F, or whether it’s some other set of factors. This problem explodes very badly as the assumed number of incoming connections increases.
References
Gasperini, M., Hill, A. J., McFaline-Figueroa, J. L., Martin, B., Kim, S., Zhang, M. D., … & Trapnell, C. (2019). A Genome-wide framework for mapping gene regulation via cellular genetic screens. Cell, 176(1-2), 377-390.
Feldman, D., Singh, A., Schmid-Burgk, J. L., Carlson, R. J., Mezger, A., Garrity, A. J., … & Blainey, P. C. (2019). Optical pooled screens in human cells. Cell, 179(3), 787-799.
