Are we using really context-specific GRN's for their nominal purpose?
[grn Warning: near-delirious rant initiating. Consider two of the major unmet needs for predicting gene function in stem cell biology:
- (optimizing cell function) What (if any) genes could be overexpressed or knocked down to improve functional outcomes in stem cell products?
- (reprogramming cell type) What genes could be overexpressed to induce a transition from cell type C to cell type D?
I want to talk about how these tasks relate to a trend in computational methods. Many papers in stem cell biology feature “context-specific regulatory networks,” meaning they make formal inferences like “Gene A regulates gene B in cell type C”. For examples, I’m talking about methods like IRENE and CellOracle.
These methods focus on predictions about reprogramming. But, on the surface they seem most useful for optimizing cell function, rather than reprogramming cell type. It’s unclear what, if anything, they should tell us about reprogramming, because reprogramming shifts the ground underneath their feet. For the network connections present only in the target cell type or only in the starting cell type, when do those appear or disappear? What if intermediate cell states have a distinctive network topology that cannot be represented as a mixture of the endpoints?
In the language of chromatin accessibility, reprogramming seems to require pioneer factors capable of opening up closed chromatin. Context-specific GRN’s seem to emphasize “settler factors” that bind in already-open chromatin regions. For example: context-specific networks inferred in fibroblasts alone, or even using a prior network derived from diverse cell type, probably would not predict that OSKM has an effect in fibroblasts. You can see a similar effect in CellOracle figure 5G, screenshotted here from the preprint (pdf). The earliest fibroblast cluster has essentially no predicted outgoing edges for the reprogramming factors used in this experiment, even though there’s clearly big effect.
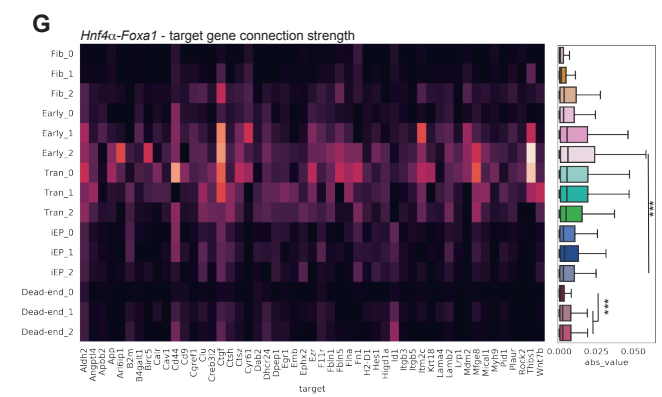
Why do these methods work well despite this glaring inconsistency? Some aspect of these methods must be doing good work without admitting to it. Maybe the factors that stabilize the target cell type are also often pioneers that can disrupt the network topology of a starting cell type. Maybe sometimes we are lucky in that way, and other times we have to give’em an extra slap. That would explain why people do this.

Peeling back a layer, this paradox is a symptom of another annoying and pervasive problem with context specific GRNs. If you want to predict FoxA2 target genes, but there is no variation in FoxA2 among your cell type of choice, then correlation != causation is the least of your worries, because you cannot even estimate the correlation alone. You could be conservative in the sense that you default to zero effect, but that’s often wrong; overexpressing FoxA2 would actually have a big effect.
Stepping back to look at the big picture, most of our knowledge and intuition about key reprogramming factors in classic cases such as MyoD and OSKM comes from studying variation across different cell types, not variation within a single cell type. Maybe computational methods should be willing to borrow information across cell types in order to make causal inferences that otherwise they cannot hope to comment on. Don’t default to 0, default to the nearest informative cell type or something like that. If you are wondering how to do this technically, consider that there is some precedent.
Closing arguments
If the regulatory network differs from C to D, we ought to ask both “How?” and “Why?” Context-specific GRN’s answer “How” in a nicely interpretable way, but:
- They are often wrong about “how” because they default to treating evidence of absence as absence of evidence. To be obtuse about it: the potential for an effect can persist in the absence of its cause.
- They give only a tautological answer to “why”: “Because it’s D and not C.”
As my mom always says, “Humph!” But could global GRN’s really do any better? Hopefully time will tell.
