Mean squared error for estimates of differential expression
Bias-variance tradeoffs for virtual cell evals are weird
Job search 2025 post-mortem and advice
The comp bio job market is scarier than usual: 2023 was a biotech bankruptcy bloodbath, 2024 was a little better, and as far as layoffs, 2025 has been worse. Academic and government sectors are also reeling from government cuts, and in the two places I have recently lived (Massachusetts and Maryland), cuts have specifically targeted large biomedical research employers (Harvard and NIH). The tech job market is pandemically weird and also not so hot. Anecdotally, there are lots of interesting job postings in comp bio, but each one might get hundreds of applicants per week, among which are many brilliant people. Some of them have training and qualifications very similar to me. I applied to about 65-70 jobs and got almost no interviews (except where I had a referral).
A recap of virtual cell releases circa June 2025
In October 2024, I twote that “something is deeply wrong” with what we now call virtual cell models. A lot has happened since then: modelers are advancing new architectures and mining new sources of information; evaluators are upping their game with deeper follow-up and an open competition; and synthetic biology wizards are building new datasets. Let’s see what’s cookin’.
Five flavors of stratified LD score regression (Part 5, Melon)
Stratified linkage disequilibrium score regression (S-LDSC) is a workhorse technique at the intersection of human genetics and functional genomics, but its exposition is heavily genetics-coded. In cell biology and functional genomics, it is less well-known and well-understood than would be optimal. This post is part of a series introducing many flavors of S-LDSC for the newcomer.
Five flavors of stratified LD score regression (Part 4, Lime)
Stratified linkage disequilibrium score regression (S-LDSC) is a workhorse technique at the intersection of human genetics and functional genomics, but its exposition is heavily genetics-coded. In cell biology and functional genomics, it is less well-known and well-understood than would be optimal. This post is part of a series introducing many flavors of S-LDSC for the newcomer.
Five flavors of stratified LD score regression (Part 3, Cranberry)
Stratified linkage disequilibrium score regression (S-LDSC) is a workhorse technique at the intersection of human genetics and functional genomics, but its exposition is heavily genetics-coded. In cell biology and functional genomics, it is less well-known and well-understood than would be optimal. This post is part of a series introducing many flavors of S-LDSC for the newcomer.
Five flavors of stratified LD score regression (Part 2, Strawberry)
Stratified linkage disequilibrium score regression (S-LDSC) is a workhorse technique at the intersection of human genetics and functional genomics, but its exposition is heavily genetics-coded. In cell biology and functional genomics, it is less well-known and well-understood than would be optimal. This post is part of a series introducing many flavors of S-LDSC for the newcomer.
Five flavors of stratified LD score regression (Part 1, Vanilla)
Linkage disequilibrium score regression (LDSC) is a workhorse technique at the intersection of quantitative genetics and functional genomics. The core use of S-LDSC is to detect and quantify functional enrichment of genetic associations; for example, “Are STAT1 binding sites enriched for genetic risk of Crohn’s Disease?” or “Are CNS-specific enhancer regions enriched for effects on psychiatric phenotypes?” or “How much disease risk is mediated by known effects on gene expression?” Unfortunately, LDSC is fairly specialized to a quantitative genetics audience, with derivations using mathematical shortcuts that may not make sense out of context. This series will give a taste of LDSC (rather, five tastes) for the newcomer, including motivation, key formulas, a summary of empirical demos, and a skippable gloss on the derivations.
Multiplexed perturbations enable massive scale ... but how big can we go?
In molecular biology, most molecules are irrelevant to most phenotypes, and it is terribly difficult to guess a mechanism a priori. This leads to bad problems.
Gene regulatory network inference is a venus fly trap for quants

Some references on genomic foundation models
I recently offered to swap bibliographies with the folks at Tabula Bio, so I put together a haphazard list of recent work on foundation models in genomics. I always want to know what are the limits of the latest data and how far we can generalize, so this discussion is a great opportunity to explore where Tabula’s interests and mine overlap. For three model classes defined by the general type of training data, here are some pointers to a sampling of existing work, plus a brief comment on where these models seem to hit a wall. Read it quick before the SOTA gets up and walks away!
Retrospection and unexpecteds, 2025 edition
Happy new year! 2025 is a round number (ends in 5) and also a square number (45^2), and it’s gonna be a BIG year for me. I have a poor memory, and I don’t often look backwards, so the calendar is giving me a useful nudge to take stock of the past 10 years.
Methods for expression forecasting under novel genetic perturbations
Forecasting expression in response to genetic perturbations is potentially extremely valuable for drug target discovery, stem cell protocol optimization, or developmental genetics. More and more methods papers are showing promising results. Let’s take a look.
Expression forecasting benchmarks
I wrote about the boom of new perturbation prediction methods. The natural predator of the methods developer is the benchmark developer, and a population boom of methods is naturally followed by a boom of benchmarks. (The usual prediction about what happens after the booms is left as an exercise to the reader.) Here are some benchmark studies that evaluate perturbation prediction methods. For each one, I will prioritize four questions:
What FDR control doesn't do
Working on transcriptome data, I use FDR control methods constantly, and I’ve run into a couple of unexpected types of situations where it’s easy to assume they will behave well… but they don’t.
Neural Network Checklist
Training neural networks is hard. Plan to explore many options. Take systematic notes. Here are some things to try when it doesn’t work at first.
Differential expression for newcomers
This post might be for you if:
You can't construct confidence intervals near a region of non-identifiability
This post will highlight Jean-Marie Dufour’s absolutely impenetrable tour de force “Some Impossibility Theorems in Econometrics With Applications to Structural and Dynamic Models”, also known by its English title, “Weak Instruments, Get Rekt.” Dufour talk about confidence intervals for a parameter $\phi$ in a situation where:
You can't test for independence conditional on a quantitative random variable.
Suppose the joint distribution of $X, Y, Z$ is continuous and the max of $X$, $Y$, and $Z$ is $M$. Suppose you want to test whether $X$ and $Y$ are independent conditional on $Z$. You want to do this with a type 1 error rate (false positive rate) controlled at 5%.
You can't estimate the mean.
Maybe this was naive of me. I thought that if you were willing to assume that the mean exists, you could probably … estimate it?
You can't satisfy three simple desiderata for a clustering algorithm.
So you want to cluster your data. And you want to use a “good” algorithm for clustering. What makes a clustering algorithm “good”?
You can't achieve optimal inference and optimal prediction simultaneously.
It’s natural to want and expect a one-stop shop. The best fitting model ought to give both the best predictions and the best inferences.
Five disturbing impossibility theorems
I remember reading, maybe in a decades-ago issue of Scientific American, that geometry and physics professors sometimes hear from cranks. These are arrogant people, working in isolation, who earnestly believe they have discovered something important. Typical examples might be trisecting an angle with a compass and straightedge in a finite number of steps or unifying physics. I have not understood the details of those examples personally, but there is a basic, recurring disconnect between the position of the crank – “The establishment has tried and failed; they do not understand” – and the position of the establishment – “We understand thoroughly; this task is proven to be impossible.”
A third miracle of modern frequentist statistics
Here’s post #3 of 4 on astonishingly general tools from modern frequentist statistics. This series highlights methods that can accomplish an incredible amount on the basis of very limited or seemingly wrong building blocks. It’s not just about the prior anymore: each of these methods works when you don’t even know the likelihood. I hope this makes you curious about what’s out there and excited to learn more stats!
A second miracle of modern frequentist statistics
Here’s post #2 of 4 on the theme of “Do 👏 not 👏 ignore 👏 the 👏 magnificent 👏 bounty 👏 of 👏 techniques 👏 from 👏 modern 👏 statistics 👏 just 👏 because 👏 someone 👏 taught 👏 you 👏 an 👏 ideology”. This series highlights tools from modern frequentist statistics, each with a distinctive advantage that nearly defies belief in terms of how much can be accomplished on the basis of very limited or seemingly wrong building blocks. It’s not just about the prior anymore: each of these methods works when you don’t even know the likelihood. I hope this makes you curious about what’s out there and excited to learn more stats!
A miracle of modern frequentist statistics
Over the past N years I’ve heard lots of smart people that I respect say things like “inside every frequentist there’s a Bayesian waiting to come out” and “Everybody who actually analyzes data uses Bayesian methods.” These people are not seeing what I’m seeing, and that’s a shame, because what I’m seeing is frankly astonishing.
A fourth miracle of modern frequentist statistics
Here’s post #4 of 4 on tools from modern frequentist statistics that a guaranteed to work despite their basis in very limited or seemingly wrong building blocks. Unlike the other miracles, you will need to specify the full likelihood for this one, but if you’ve only ever been exposed to Bayesian takes on variational inference, it may open your mind to interesting new possibilities, or at least alert you to an issue you might not have thought about yet.
Are we using really context-specific GRN's for their nominal purpose?
Warning: near-delirious rant initiating. Consider two of the major unmet needs for predicting gene function in stem cell biology:
Learning to learn about epigenetics
I just finished teaching a one-credit engineering class (part of JHU’s HEART). It was an opportunity for students and me to indulge our curiosity about a really neat topic: namely, epigenetics.
Learning to learn about epigenetics (example project)
This is an example of a completed project from my class on epigenetics.
Learning to learn about epigenetics (project description)
This is a description of the research project from my class on epigenetics.
Lazy matrix evaluation saves RAM in common analyses
MatrixLazyEval is an R package for “lazy evaluation” of matrices, which can help save time and memory by being smarter about common tasks like:
kniterati lets you develop R packages in R markdown
For a while, I experimented with writing all of my R packages in R markdown. In order to facilitate this, I built kniterati, which knits all the Rmd files into R files prior to the usual devtools workflow. Check it out at https://github.com/ekernf01/kniterati.
freezr keeps results with the code that produced them
Doing single-cell RNA analysis for the Maehr Lab, it was a constant struggle to keep everything organized. The nightmare scenario would be to get a useful or interesting result during an hour of intensive experimentation and re-writing of code, then later discover that the code to produce that result no longer exists. I wanted to interact with the data, not be interrupted constantly to commit microscopic changes, so I wrote freezr, which saves code, plots, and console output to a designated folder just as the same code runs. Check it out at https://github.com/ekernf01/freezr.
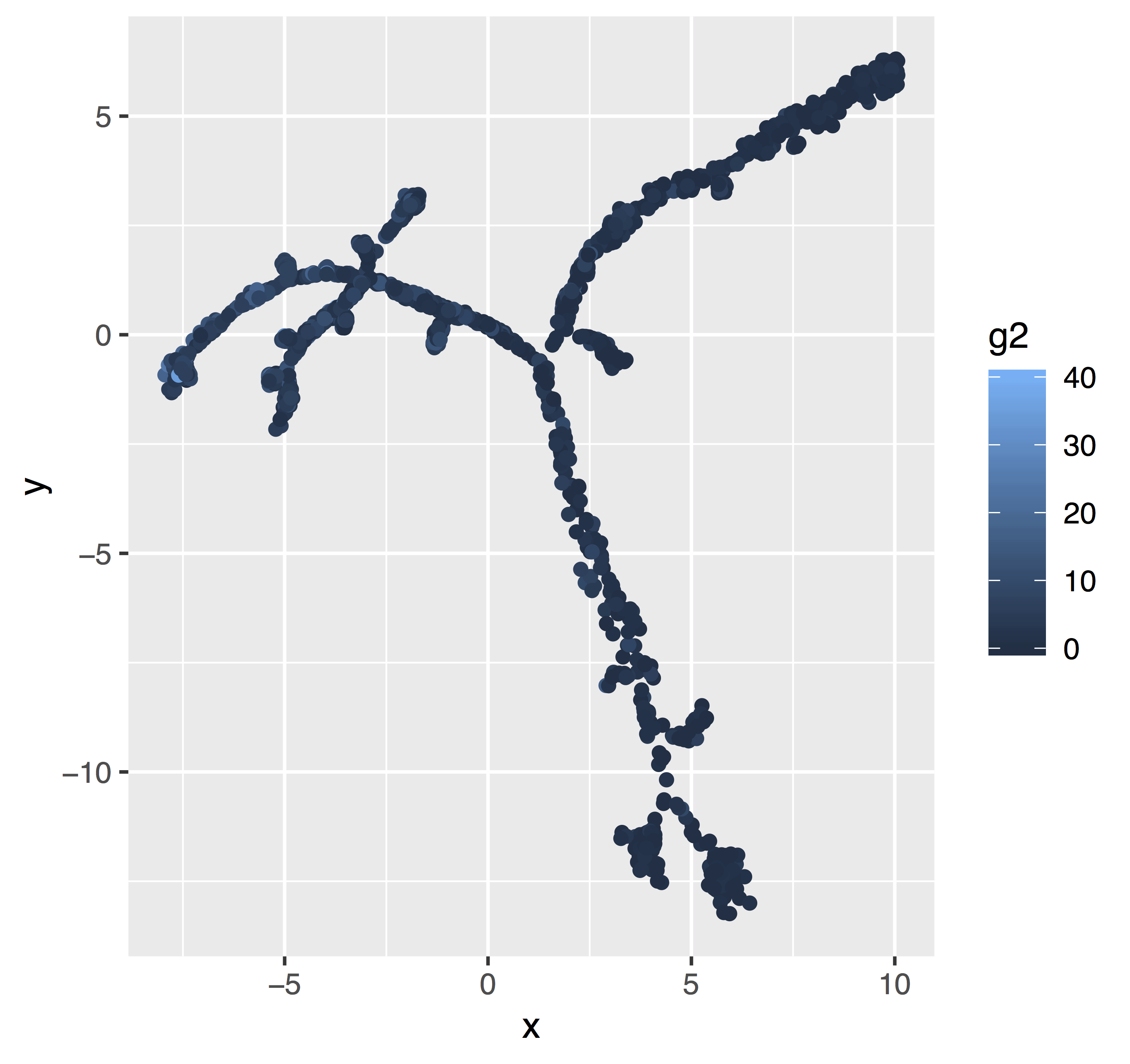
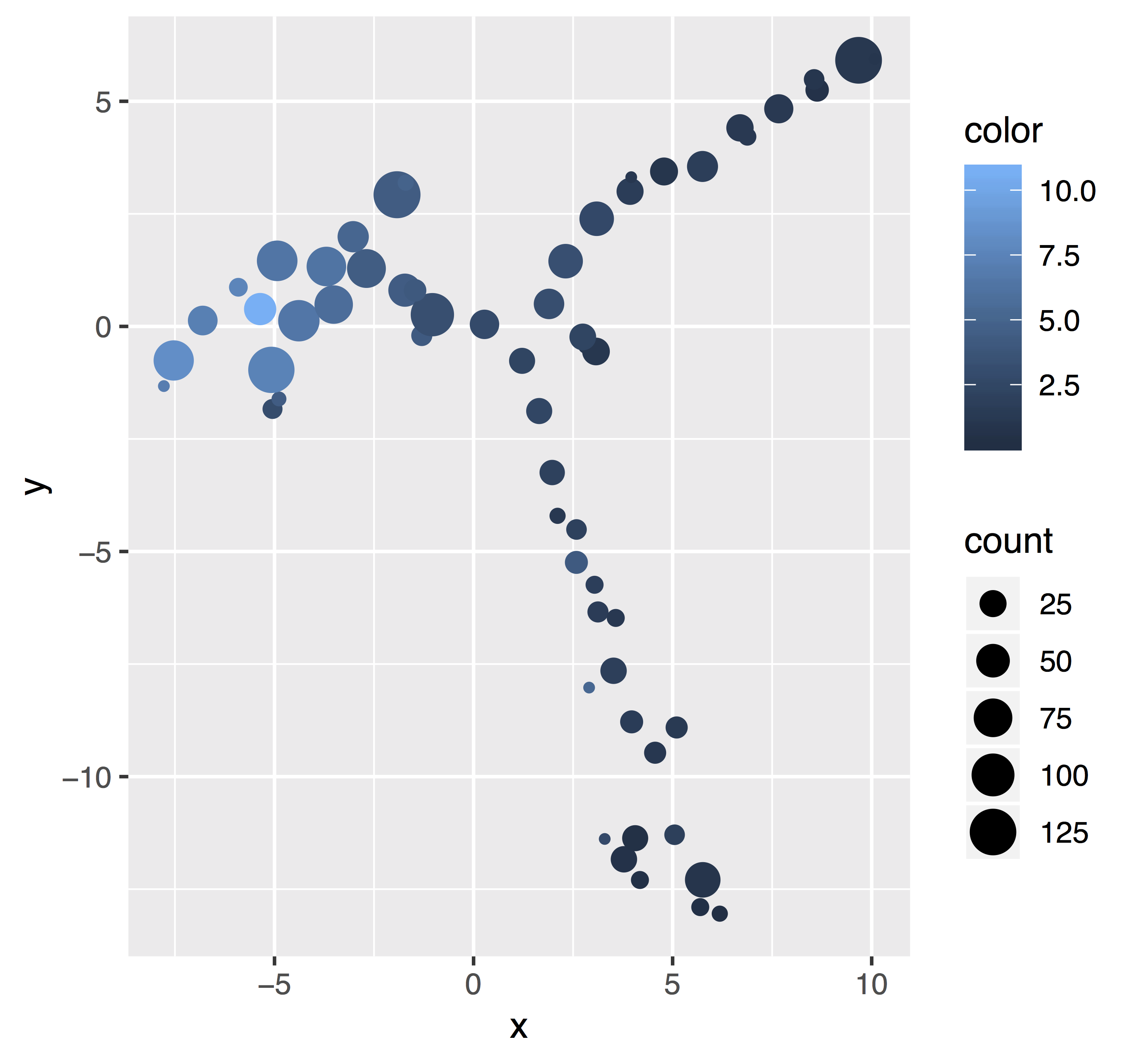
Bubble plots resolve overplotting
Kalman Filter Cheat Sheet
To help me understand Kalman filtering while studying for quals, this cheat sheet condenses and complements the explanation of the Kalman filter in Bishop PRML (pdf) section 13.3. I wrote this as if I were about to implement it. (I didn’t: there’s already an open-source implementation, pykalman, including all the functionality discussed herein.) I’m having trouble with math typesetting on the web, so here’s the markdown and pdf of this post.
Experiments with one control and $k$ treatments have highest power when the control arm is $\sqrt k$ times bigger than each individual treatment arm
When we screened transcription factors influencing endoderm differentiation (post, paper), we ran into an interesting design problem. The experiment had 49 different treatment conditions and one control. Treatments were not allowed to overlap. We were able to measure outcomes in a fixed number of cells – as it turned out, about 16,000. What is the optimal proportion of cells to use as controls?
Network modeling and the Connectivity Map
Here’s a puzzle for you: why is nobody using CMAP for network modeling in stem cell biology?
Simple isoform abundance estimation
Sorry, this one’s not ready yet!
Inferring developmental signals from RNA-seq data
Back in the Maehr lab, any given experiment that my coworkers ran was expensive and hard. Our most common measurements – flow cytometry, imaging, and qPCR – could take hours or a day or two. Making a batch of virus containing CRISPR guide RNAs could take days or a week or two. Running a directed differentiation could take weeks or a month or two, especially if the cells died. Making a cell line could take months. So if they are going to design an screen, they really want a good chance to find something in there that works for whatever result they are trying to obtain.
Problems in causal modeling of transcriptional regulation
Earlier, I discussed opportunities and ambitions in modeling the human transcriptome and its protein and DNA counterparts. I also promised a discussion of the many statistical issues that arise. To begin that discussion, I am trying to identify statistical issues and position them into some type of structure.
Coping with missing data in biological network modeling
This is not a standalone post. Check out the intro to this series. This particular post is about the number one limitation in causal network inference: missing layers. In the most common type of experiment, we measure gene activity by sampling RNA transcripts, reading them out, and counting them. We can only guess at:
Intro to gene regulatory network post series
The mystery of life
Enhancer integration for network modeling
In the intro, I described a three-part scheme to unravel the mystery of multicellular life. As part of that, I talked about how I mostly am trying to predict RNA levels these days. But, we know that important parts of the human regulatory network are contained in other types of molecules: for example, of all mutations related to autoimmunity, 90% are not in a coding region. This post discusses a class of gene-like entities called enhancers that have recently emerged as an interesting and potentially useful counterpart to genes. Teams are beginning to catalog enhancers and figure out how they help control cell state. This post will survey how that’s being done and will convey one important current question: how do we best connect each enhancer with the gene(s) it helps control?
Data resources for gene regulatory network modeling
This is not a standalone post – it’s just a rough list of datasets that could be useful for regulatory network models. Check out the intro to this series for more context.
DNA ==> RNA ==> Protein
This post is part of the GRN series. Check out the intro.
How much data? A cool perspective from 1999 (gene regulatory network series)
In the intro, I discussed the puzzle of multicellular life: many cell types, one genome. I also followed up by discussing the many statistical issues that arise. There’s a very cool paper from 1999 that brings a lot of clarity to this situation, and here I have space to dig into it a bit.
Favorite machine learning papers (resnets)
I’ve been reading recently about debugging methods, starting with this post by Julia Evans. All the links I followed from that point have something in common: the scientific method. Evans quotes @act_gardner’s summary:
Contra Rubin et al on genetic interations
In the Maehr lab’s regular journal club, we recently discussed a Cell paper from the research groups of Howard Chang, Will Greenleaf, and Paul Khavari (henceforth “Rubin et al.”).
I don't want to be afraid of neural networks anymore
Neural networks are exploding through AI at the same pace as single-cell technology in genomics and GWAS consortia in genetics – that is to say, very quickly. For one example, ImageNet error rates decreased tenfold from 2010 to 2017 (source), but people are also now using neural networks to play StarCraft at a superhuman level, synthesize and manipulate photorealistic images of faces, and yes, also to analyze single-cell genomics data. This seems like a class of models I should consider learning to work with.
Definitive endoderm screen
The Maehr Lab recently published another paper! Hooray! I want to recap it with a quick summary and some technical things I learned on the project.
Wellness rewards programs allow discrimination (technical appendix)
This is not a stand-alone post. It is a technical appendix for an upcoming post, which is (EDIT) now published here on the blog of the peer-reviewed journal Medical Care!
We want R functions for gradients and hessians
The programming language R typically offers functions prefixed by d, p, q, and r for pdf, cdf, inverse cdf, and sampling. These are useful for fitting statistical models – especially MCMC since you’re usually sampling and computing densities. You can even get the log density from the d function with an extra argument.
What I wish I knew two years ago, or, best practices for command-line tools in bioinformatics
I work in a small lab. The number of bioinformaticians hovers around 1 to 1.5. We prioritize interaction with the data, so we do not spend the effort to implement things from scratch unless we absolutely need to. We start with what’s out there and adapt it as necessary. That means I have installed, used, adapted, or repurposed many shapes or sizes of bioinformatics tools. In terms of usefulness, they run the gamut from “I deeply regret installing this” to “Can I have your autograph?”. Some patterns emerge distinguishing those that are most pleasant to work with, and that’s the topic of today’s post.
Thymus Atlas
Why study the thymus? The eponymous “T cells” of the immune system:
The curious case of the missing T cell receptor transcripts, part 3
This is part 3 of a three-part post on T-cell receptors & RNA data. Here are parts 1 (intro / summary) and 2 (technical details).
The curious case of the missing T cell receptor transcripts, part 2
This is part 2 of a three-part post on T-cell receptors & RNA data. Here are parts 1 (intro / summary) and 3 (bonus material).
The curious case of the missing T cell receptor transcripts
This is part 1 of a three-part post on T cell receptors & RNA data. Here are parts 2 (technical details) and 3 (bonus material). Also check out WAT3R, a custom-built analysis pipeline for TCR + scRNA data. I wasn’t involved in developing WAT3R, but it looks like a really nice tool, and much more modern than this post you’re reading.
About the Maehr lab
Addendum 2020 August 15: I worked in the Maehr lab for almost 4 years. I am now excited to be entering the Ph.D. program in the Johns Hopkins BME Department.
Configuring pipelining tools for LSF
Many (all?) bioinformatics groups use cloud or cluster computing to handle grunt work such as sequence alignment. They use scheduling systems such as Sun Grid Engine and LSF to submit jobs to the cluster. But, it’s becoming more common to use one of many modern pipelining tools. These pipelining tools abstract away the details of job submission, getting rid of boilerplate that would otherwise appear every time you build a pipeline.
Tracer-seq does not infer lineage trees, per se
This is the fully detailed version of this post about TracerSeq. If you find it daunting, boring, or overly technical, check out the short version first.
Tracer-seq does not infer lineage trees, per se
This is a short version of the full post about TracerSeq.
Hi Mom! Hello world!
Hello world. Stay tuned for initial posts, coming up!
